An identification key, also known as a taxonomic key, is a
useful tool for identifying unknown organisms. Keys are constructed so that
the user is presented with relevant information in a structured form. This
allows to skip over the many species that do not possess certain characters
and is supposed to lead the user to a correct identification with a minimum
number steps.
Keys that are based on subsequent choices between two character states are
called dichotomous keys. Such keys are written using pairs of contrasting
characteristics (known as couplets), where the choice of one character state
leads to another couplet until the organism is identified.

Figure 1. Typical identification key, here for the species of the genus Epinephelus (Heemstra and Randall 1990).
Most keys are paper-based (Figure 1) and identification
using this method is a tedious and time consuming work. For example, choice
of a wrong character state will lead to a string of couplets that appear
more and more unrelated to the organism under study. Going backwards is
typically not supported by printed keys, and as a result, the users often
have to start all-over.
Computer based identification keys have more options and typically support
back-tracking and provide pictures of excluded species as well as of those
that are still under consideration. This considerably facilitates the
identification process.
The identification keys in FishBase come with two interfaces: a simple but
efficient ‘FB keys’ interface, which shows the couplets extracted from
printed keys and adds back-tracking and photos of species; and the LucID
Phoenix identification software developed by CSIRO, Australia, which uses
the same couplets but adds additional features such as list of species
exclude and still considered (Figure 2 and 3); this software requires
download of a Java applet.
With the FB keys interface, users are presented with the complete list of
couplets. Choosing a character state by clicking its number moves the
respective next couplet to the top of the page. Links and photos or drawings
are provided to show the described species or the characters being
mentioned. The number of the respective previous couplet is shown,
supporting step-by-step back-tracking. In essence, the FB keys
identification interface is still similar to a paper-based identification
key, but already easier to use.
The Lucid Phoenix identification system is a special software written in
Java. It provides a user-friendly interface that displays all relevant
information in four side-by-side windows. The upper left window shows the
couplet under consideration plus explanatory drawings, if available. The
upper right window shows thumbnail photos of species that are still under
consideration. The lower left window shows a list of the character states
that have been selected, and which on click can be revisited. The lower
right window shows thumbnail pictures of species that have been excluded. As
CSIRO puts it: “Lucid Phoenix is a computer based dichotomous or pathway key
Builder and Player that enables traditional paper based identification keys
to be published on the Internet or CD. Phoenix keys are interactive, can be
enhanced with multimedia, and delivered across the Internet seamlessly”
(http://www.lucidcentral.org/phoenix/).

Figure 2. FB Keys interface
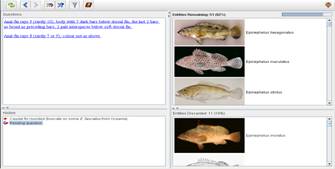
Figure 3. Lucid interface
The IDENTIFICATION KEYS Tables
Information that is used by the FB Keys and Lucid Phoenix interfaces is stored in the KEYS and the KEYSQUESTIONS tables. Figure 4 shows the encoder interface to these tables. The KEYS table contains general information about the keys, which can refer to Orders, Families, Genus or Species. The table includes the following fields:
Family: Code number of the Family which the key refers to; blank if more than one Family is covered by the key.
FAO Area: Code number of FAO Area which is covered by the key; blank if more than one FAO area is covered.
Country: 3-digit UN code of country or area which is covered by the key; blank if more than one country is covered.
Ecosystem: Code number of the ecosystem which is covered by the key; blank if more than one ecosystem is covered.
Description: Gives a short description of the key, typically including taxonomic group and area,; often identical with the title of the key as published.
Remarks: Additional remarks about the key.
LarvalBase: A Yes/No field indicating whether the key refers to eggs or larvae.
Pictures: File names of images presenting terms and measurements being used in the key.
The KEYQUESTIONS table contains the following fields:
KeyCode: Internal code number of keys.
Couplet Number: Identifier of couplet (integer) and questions (a, b, c), such as 3a.
Question: Contains one question about a character state as part of a couplet.
Next couplet: Number of the next couplet if the answer is yes, blank if it is a terminal question.
Previous couplet: Number of the previous couplet, blank if no previous couplet or question exist.
Order, Family, Genus, Species: Indication of taxon code or name if the respective question is a terminal one.
PicName: File name of picture showing details of the character in question.
Next Key: Code number of the subsequent key, if, for example, a key to Orders is connected to a key to Families.
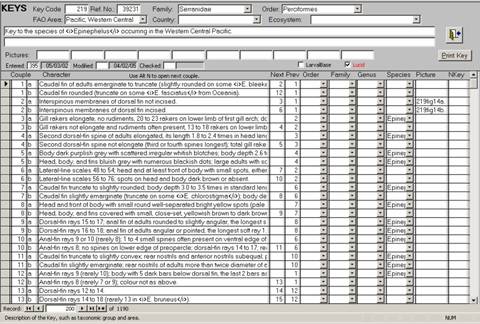
Figure 4.
Encoder interface to the KEYS and KEYQUESTIONS tables.
How to get there
Internet
We used published keys from a variety of books and journals. Important sources were FAO Species Identification Guides for Fishery Purposes (e.g. Carpenter 2002), FAO Species Catalogues (e.g. Heemstra and Randall 1990), and keys published in the Fauna Sinica series, (e.g. Chen et al. 1998, Yue et al. 2000, Chen 2002).
Chen, S.. 2002. Fauna Sinica. Ostichthyes. Myctophiformes, Cetomimiformes, Osteoglossiformes. Science Press, Beijing. 349 p.
Chen, Y.-Y. and et al. (Eds). 1998. Fauna Sinica. Osteichthyes. Cypriniformes II. Science Press, Beijing, 531p.
Heemstra, P.C. and J.E. Randall. 1990. FAO species catalogue. Vol. 16. Groupers of the world (family Serranidae, subfamily Epinephelinae). An annotated and illustrated catalogue of the grouper, rockcod, hind, coral grouper and lyretail species known to date. FAO Fish. Synop. 125(16):382 p.
K.E. Carpenter (ed.). 2002. FAO species identification guide for fishery purposes. The living marine resources of the Western Central Atlantic. Vol. 1: Introduction, molluscs, crustaceans, hagfishes, sharks, batoid fishes, and chimaeras.
P. Yue et al. (Eds). 2000. Fauna Sinica. Osteichthyes. Cypriniformes III. Science Press. Beijing. 1-661.
Sheryl Yap and Rainer Froese, October 2005